Prof Brian Kobilka’s group reported structural insights into the process of GPCR-G Protein complex formation
On 9th May 2019, Xiangyu Liu and Xinyu Xu in Professor Brian Kobilka’s group in the School of Medicine, Tsinghua University, published an article entitled “Structural Insights into the Process of GPCR-G Protein Complex Formation” in the journal Cell. The paper reported an active β2AR structure obtained by fusion with the C terminus peptide of the Gαs protein (GsCT) and the structure of GDP bound Gs heterotrimer (GsGDP). These structures provide evidence for an alternate interaction between the β2AR and Gs that may represent an intermediate that contributes to Gs coupling specificity. In the same issue of Cell, Professor Brian Kobilka’s group in the School of Medicine, Stanford University, published a paper entitled “Assembly of a GPCR-G protein complex”, which reports their efforts to investigate the dynamic process for GPCR-G protein complex formation using time-resolved mass spectrometry. These two papers support each other and reveal the molecular mechanism of GPCR-G Protein complex formation from different aspects.
G protein-coupled receptors (GPCRs) are the largest family of membrane receptors with more than 800 family members in the human genome. GPCRs play important roles in physiology by mediating the senses of sight, smell, taste and the responses to hormones, and neurotransmitters. They are the targets of approximately 30% of clinical drugs. The activated receptors bind to a diverse set of downstream signaling proteins including G proteins, GRKs and arrestins. An agonist bound GPCR first forms complex with guanosine diphosphate (GDP) bound G protein (GPCR-GGDP), triggers the release of GDP from Gα, leads to the formation of a nucleotide-free complex (GPCR-Gempty). Then guanosine triphosphate (GTP) binds to G protein and dissociates the complex to allow different subunits to activate downstream signaling pathways (Figure 1).
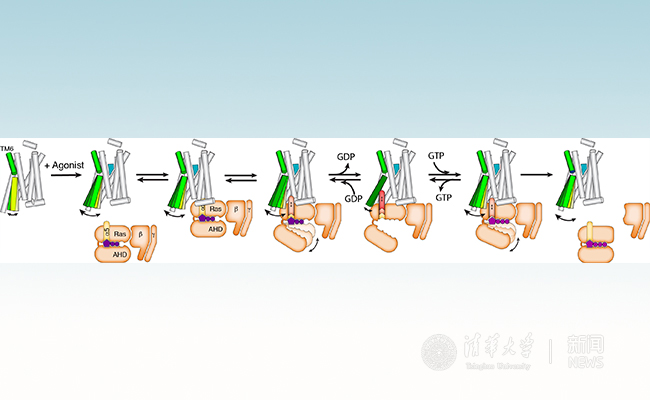
Figure 1. Process of GPCR-G Protein Complex Formation
The β2 adrenergic receptor (β2AR) has been a model system for GPCRs studies. Professor Brian Kobilka’s group at Stanford University determined the inactive state β2AR structure in 2007, which was the first crystal structure of a hormone and neurotransmitter receptor. The β2AR-Gsempty complex structure was subsequently determined by X-ray crystallography in 2011, which was the first crystal structure of an intact GPCR-G protein complex. Recently, several family A and family B GPCR structures in complex with either Gs or Gi/o proteins were determined by single-particle cryo-electron microscopy (cryo-EM). All of these complexes were captured the nucleotide-free state (GPCR-Gempty). Unfortunately, these GPCR-Gempty complex structures do not provide a clear explanation for G protein coupling specificity. Evidence from several sources suggests the existence of a transient complex between the GPCR and GDP-bound G protein (GPCR-GGDP) that may represent an intermediate on the way to the formation of GPCR-Gempty and may contribute to coupling specificity. However no structural information was available for the GPCR-GGDP complex.
Professor Brian Kobilka’s group has observed that agonists alone do not fully stabilize the active state of the β2AR. The active conformation of β2AR required both extracellular agonists and intercellular binders such as G proteins or G protein mimic nanobodies. In an effort to develop a general applicable method to stabilize GPCRs in active conformation, the Kobilka lab at Tsinghua developed a protein engineering strategy by inserting the last 14 residues from the C terminus of Gαs (GsCT) between TM5 and TM6 of the β2AR and introducing a disulfide bond to stabilize the interaction between the β2AR and GsCT. The construct showed increased agonist affinity (a sign of active β2AR conformation) and was named as β2AR-T4L-GsCT-CC. Finally, a β2AR-T4L-GsCT-CC structure was obtained at 3.7 A resolution by crystallography and the β2AR indeed adopts active conformation in the structure. Interestingly, the interactions between β2AR and GsCT observed in β2AR-T4L-GsCT-CC structure are different from those in the β2AR- Gsempty complex (PDB:3SN6). Y391Gαs and H387Gαs, which interact with the β2AR in the β2AR-Gsempty complex, are facing the solvent in the β2AR-T4L-GsCT-CC structure. As a consequence of the rotation of the helix, E392Gαs and R389Gαs, which face the solvent in the β2AR-Gsempty complex, are interacting with the core of the β2AR (Figure 2). MD simulations suggest that the interactions in the β2AR-T4L-GsCT-CC structure are nearly as stable as those in the β2AR-Gsempty complex structure.
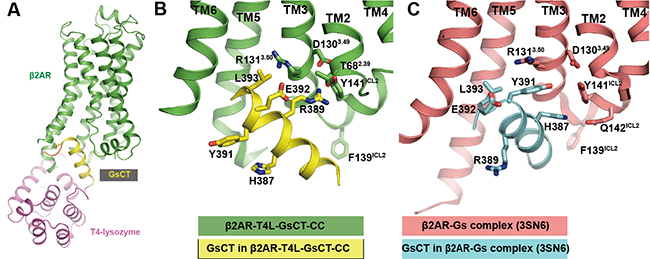
Figure 2. Overall β2AR-T4L-GsCT-CC structure and alternative interaction between GsCT and β2AR compared to the β2AR-Gs complex structure (3SN6).
They then solved the GDP-bound Gs heterotrimer structure (GsGDP) at 2.8 A resolution. In the GsGDP structure, Y391Gαs and H387Gαs form intermolecular interactions with other residues in Gαs while E392Gαs and R389Gαs are exposed to the protein surface, which suggests E392Gαs and R389Gαs are more likely to involve in the initial interaction with the β2AR. They subsequently confirmed the important roles of E392Gαs and R389Gαs in efficiently β2AR-Gs complex formation by mutagenesis studies. Finally they proposed a model of the dynamic process of the GPCR-G protein complex formation from separated β2AR and GsGDP to the β2AR-GsGDP complex and eventually to the β2AR-Gsempty complex (Figure 3). They also provided evidence that this intermediate state complex contributes to the G protein coupling specificity.
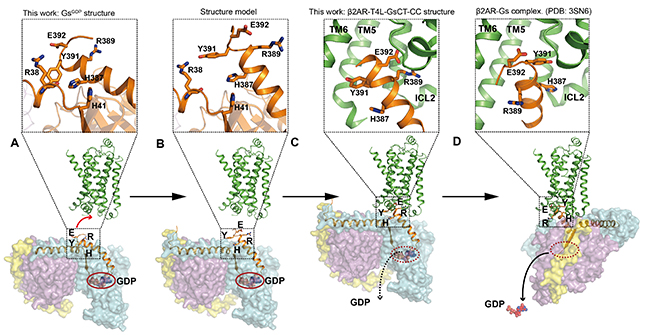
Figure 3.Proposed Process of GPCR-G Protein Complex Formation
This work is the result of a collaboration between the Kobilka Lab at Tsinghua University and Stanford University, and Professor Peter Hildebrand’s group at Charite Medical University Berlin and University of Leipzig, as well as Dr. Jesper Mathiesen from University of Copenhagen. Professor Brian Kobilka and Dr. Xiangyu Liu from School of Medicine, Tsinghua University are the co-corresponding authors; Dr. Xiangyu Liu, Xinyu Xu (Tsinghua University) and Dr. Daniel Hilger (Stanford University) are the co-first authors. Dr. Hongtao Liu, Dr. Xiaoou Sun from Tsinghua University and Dr. Yang Du from Stanford University (current address: Kobilka Institute of Innovation Drug Discovery (KIIDD), the Chinese University of Hongkong, Shenzhen) also contributed to the work. Dr. Kunio Hirata from Spring-8 synchrotron radiation facility performed automatic data collection for the project. The work was supported by the Beijing Innovation Center for Structural Biology.
The original link:
https://www.cell.com/cell/fulltext/S0092-8674(19)30440-4
(From School of Medicine)

